The physical properties of materials, ranging from atoms to proteins, are determined by their underlying structure. Our research focuses on leveraging advanced electron microscopy technology to elucidate the atomistic structure of novel materials. By understanding these structures, we aim to uncover opportunities in diverse applications such as quantum computing, renewable energy, and drug discovery.
(1) Structure and properties of two-dimensional (2D) materials
We employ a combination of traditional scanning transmission electron microscopy (STEM) techniques and state-of-the-art detectors to investigate the intricate atomic structure and broader physical properties of emerging two-dimensional (2D) materials. Our ultimate objective is to achieve precise control over the structural arrangement, electronic properties, and quantum effects of these materials, with the aim of unlocking their potential for applications in low-power electronics and quantum computing.
- Chuqiao Shi, Nannan Mao, Kena Zhang, Tianyi Zhang, Ming-Hui Chiu, Kenna Ashen, Bo Wang, Xiuyu Tang, Galio Guo, Shiming Lei, Longqing Chen, Ye Cao, Xiaofeng Qian, Jing Kong, Yimo Han “Domain-dependent Strain and Stacking in Two-dimensional van der Waals Ferroelectrics” Nature Communications, 14, 7168 (2023).
- Y. Han*, M. Li*, G. Jung*, M. A. Marsalis, Z. Qin, M. J. Buehler, L. Li, D. A. Muller, “Sub-nanometer Channels Embedded in Two-Dimensional Materials,” Nature Materials, 17, 129-133 (2018).
- S. Xie, L. Tu*, Y. Han*, L. Huang, K. Kang, K.U. Lao, P. Poddar, D. A. Muller, R.A. DiStasio Jr., J. Park, “Coherent Atomically-thin Superlattices with Engineered Strain,” Science, 359, 1131-1136 (2018).
(2) Characterization of soft materials in liquid environments.
Our research employs cutting-edge cryogenic electron microscopy (cryo-EM) and tomography (cryo-ET) techniques to obtain high-resolution insights into the dynamic and functional behavior of biomolecules in their native environment. Additionally, we also explore the interactions and interfaces between biological systems and nanomaterials, shedding light on their mutual influence and potential applications in fields such as drug delivery, bioimaging, and nanomedicine.
- Heng Ni, Xiao Fan, Feng Zhou, Galio Guo, Jae Young Lee, Ned Seeman, Do-Nyun Kim, Nan Yao*, Paul Chaikin*, Yimo Han*, “Direct Visualization of Floppy 2D DNA Origami using Cryo-EM,” iScience, 25, 6, 104373 (2022).
- Y. Han, X. Fan, H. Wang, F. Zhao, C.G. Tully, J. Kong, N. Yao, N. Yan, “High Yield Mono-layer Graphene Grids for Near-Atomic Resolution Cryo-Electron Microscopy,” PNAS, 117, 1009-1014 (2020).
- L. Wang*, H. Qian*, Y. Nian*, Y. Han*, Z. Ren, H. Zhang, L. Hu, B.V.V. Prasad, A.Laganowsky, N. Yan, M. Zhou, “Structure and mechanism of human diacylglycerol acyltransferase 1,” Nature, 581, 329-332 (2020).
(3) Computational electron microscopy and machine learning
We develop cutting-edge tools and techniques to achieve higher resolution, enhanced precision, and larger-scale mapping for the characterization of materials and biomolecules. We also utilize machine learning methods to automate data processing and parameter tuning, enabling more efficient and accurate analysis of complex datasets.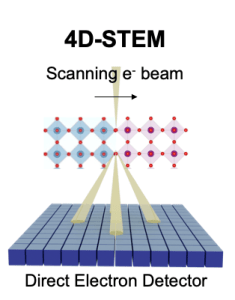
- Chuqiao Shi, Michael C. Cao, Sara M. Rehn, Sang-Hoon Bae, Jeehwan Kim, Matthew R. Jones, David A. Muller, Yimo Han*, “Uncovering Material Deformations via Machine Learning Combined with Four-Dimensional Scanning Transmission Electron Microscopy,” npj Computational Materials, 8, 114 (2022).
- Z. Chen, M. Odstrcil, Y. Jiang, Y. Han, M.-H. Chiu, L.-J. Li, D.A. Muller, “Mixed-state Electron Ptychography Enables Sub-angstrom Resolution Imaging with Picometer Precision at Low Dose,” Nature Communications, 11, 1-10 (2020)
- Y. Jiang*, Z. Chen*, Y. Han, P. Deb, H. Gao, S. Xie, P. Purohit, M.W. Tate, J. Park, S.M. Gruner, V. Elser, D.A. Muller, “Electron ptychography of 2D materials to deep sub-ångström resolution,” Nature, 559, 343-349 (2018).